Research
Overview

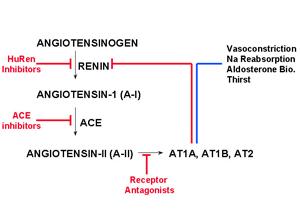
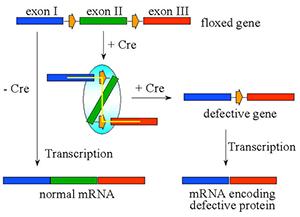
Renin-angiotensin System cre-loxP Recombinase System
My laboratory is primarily interested in investigating the regulation of genes involved in cardiovascular homeostasis and in creating new transgenic and knockout models of cardiovascular disease that will allow us to explore how these genes regulate blood pressure. We use state-of-the-art molecular and physiological approaches to these problems including the extensive use of genetic manipulation including transgenesis, gene targeting and viral gene delivery. For the past 12 years, we have been investigating the mechanisms by the renin-angiotensin system (RAS) regulates blood pressure in normal and pathological states. We have developed novel models with which to explore the importance of RAS gene expression in the kidney and brain. We also make extensive use of both BAC-mediated transgenesis and the cre-loxP recombinase system. More recently, we have been exploring the mechanism by which the PPARg transcription factor regulates blood pressure and vascular tone. This new project is making extensive use of viral gene delivery, transgenesis, dominant negative mutants, microarrays and computational biology. Additional details on projects in the Sigmund lab is available through individual links.
Tissue Renin-Angiotensin Systems in Blood Pressure Regulation
An intrinsic tissue renin-angiotensin system (RAS) is defined as a tissue-specific system with the potential for the local generation and action of Ang-II. We and others hypothesize that this local generation of Ang-II may provide a level of local control independent of the circulating (or endocrine) system. It has been shown that the kidney and brain both express each RAS gene mRNA and contains each RAS product. Therefore, the kidney and brain may have the capability of generating Ang-II from AGT released locally from renal proximal convoluted tubules (PCT) or astrocytes, respectively. Before the inception of this project, the concept that intrinsic tissue renin systems existed was highly controversial because it was not experimentally feasible to physiologically separate the effects of tissue RAS from the endocrine RAS. We have since provided the first convincing evidence in support of the blood pressure regulatory function of a tissue RAS in the kidney. This was accomplished with the use of a transgenic model in which angiotensinogen (AGT) was specifically targeted to renal PCT cells and studies demonstrating that transgenic mice expressing RAS components specifically within the kidney exhibited chronic hypertension without changes in circulating Ang-II. Importantly, we recently demonstrated that the cre-loxP recombinase system could be used as an effective tool to generate a tissue-specific knockout of RAS genes to experimentally dissect tissue RAS. We will examine the overall hypothesis that intrinsic tissue renin-angiotensin systems play an integral role in the regulation of blood pressure and may participate in the development or maintenance of hypertension. We will focus on the specific aims: 1) we will test the hypothesis that the intra-renal RAS plays a critical role in the regulation of blood pressure and renal function, and when specifically ablated via the cre-loxP recombinase system will reduce blood pressure in a model of Ang-II-dependent hypertension, lower basal blood pressure in normotensive mice, and alter renal function, and 2) we will explore the hypothesis that an intrinsic RAS in the brain, derived from locally synthesized AGT (and therefore Ang-II), plays an important role in the regulation of basal blood pressure, and when specifically ablated will lower blood pressure in a model of Ang-II-dependent hypertension by altering central effector mechanisms.
Molecular Mechanisms Regulating Renin Gene Expression
For the past 10 years we have been examining the regulation of the gene encoding a protein (renin) central to the regulation of blood pressure and water and electrolyte homeostasis. We have identified a strong enhancer of transcription 2.6 kb upstream of the mouse renin gene and its homolog 13.0 kb upstream of the human renin gene. This enhancer is a complex regulatory element consisting of the binding sites for at least 7 different transcription factors including NF-Y, RARa, RXRa, CREM, CREB, USF-1 and USF-2. These transcription factors serve to both up-regulate and down-regulate renin expression in response to physiological cues. Using yeast one-hybrid analysis, we recently determined that Ear2, an orphan member of the nuclear hormone super-family, is a strong negative regulator of renin expression. It represses transcription by binding to the same site as RARa on the enhancer and may therefore compete with it for binding. We have also generated a number of transgenic mouse models to examine the regulation of renin transcription. The most important model consists of a 160 kb P1 artificial chromosome (called PAC160) construct which contains the renin gene as well as the upstream PEPP3 and Kiss1 genes and the downstream FLJ10761 and Sox13 genes. The regulation of renin transcription from this construct is exquisitely tight, responding exactly as anticipated to physiological signals which normally regulate the renin gene. We are currently performing mutagenesis of each of the transcription factor binding sites in this PAC construct to assess their importance in tissue- and cell-specific expression of renin, its response to hormonal stimulation and its response to physiological signals. We are also determining if the enhancer influences the transcription of the nearby upstream and downstream genes.
Physiological Significance of Genetic Variation in the Human Genome
We are testing the overall hypothesis that allelic variation in a gene(s) causes a change in its function rendering it a hypertension or atherosclerosis -causing or -susceptibility gene, and that defined manipulation of the mouse genome can be used to test the physiological significance of genetic variants in human genes, and its interaction with environmental and genetic stressors. The studies proposed herein will provide mechanistic studies on the biological consequences of variations of these genes and on the interaction of genetic and non-genetic factors that modulate the susceptibility to disease. We propose to examine two common allelic or haplotypic variants of each of six genes by generating mouse models in which identical genes differing only at the variant positions are specifically targeted to the mouse genome in a way which will allow direct comparison among genetic variants. In each case we will compare the effects of allelic variation on baseline blood pressure, vascular function and the genesis of atherosclerosis. Moreover, the contribution of each allelic variant on vascular function, hypertension and atherogenesis will be compared in mice induced to be hypertensive either genetically or experimentally, in mice induced to become atherosclerotic, and in mice under environmental stressors such as a high sodium diet, high fat diet or with other risk factors such as obesity and atherosclerosis. To accomplish this goal we will: 1) Generate “knockin” mice in which each gene variant is specifically targeted in a single copy to a single defined locus in the mouse genome, and where appropriate breed each gene variant to a genetic background deficient in endogenous mouse gene of interest, 2) Compare baseline blood pressure, vascular function, and atherogenesis in each set of allelic models, 3) Compare blood pressure, vascular function and atherogenesis in mice made genetically or experimentally hypertensive, in mice made genetically atherosclerotic or obese or challenged by other CVD risk factors such as a high salt or high fat diet. These aims will allow us to assess the physiological importance of allelic variation in six genes implicated as risk factors in the pathogenesis of human essential hypertension or atherosclerosis.
Role of PPARg in Hypertension and Vascular Function
Peroxisome proliferator activated receptors (PPAR’s) are ligand activated transcription factors which have a pleiotropic role in many physiological processes. PPARg is the molecular target of the thiazolidinediones class of drugs which are used to treat patients with non-insulin dependent diabetes mellitus (NIDDM). Endothelial dysfunction, which develops in patients that are diabetic or chronically hypertensive, is thought to contribute to the progression of carotid artery disease, cerebral vascular dysfunction and stroke. PPARg is expressed in vascular endothelium and smooth muscle and therefore is a potentially important factor in the regulation of vascular function and blood pressure. PPARg has been reported to inhibit responses to vasoconstrictors such as endothelin, stimulate the release of vasodilators, and increase expression of CuZn-SOD in vascular muscle and endothelium. Additionally, treatment with thiazolidinediones has been reported to lower blood pressure in patients with NIDDM. Moreover, patients carrying dominant negative mutations in PPARg exhibit early onset type II diabetes and hypertension. Current data suggests that PPARg exerts a protective effect in the vessel wall. We hypothesize that PPARg plays an important role in the regulation of vascular function and blood pressure. We will test this hypothesis using transgenic and viral transfer technologies, in order to gain new insight into PPARg’s physiological and pathophysiological role in the carotid artery and cerebral circulation. To test this hypothesis, we will 1) test whether PPARg activation improves endothelial function and lowers blood pressure in a transgenic mouse model with established endothelial dysfunction and hypertension, 2) use adenoviruses over-expressing wildtype and dominant negative mutations of PPARg in carotid arteries from normotensive and hypertensive mice to test whether they can alter endothelial function, and 3) generate novel transgenic mice with expression of the wild-type and dominant negative mutants of PPARg targeted specifically to vascular muscle and endothelial cells using cell-specific promoters. We will characterize vascular function in these models to gain a better understanding of PPARg role in the vasculature both under the normal and diseased conditions. We will also use microarrays and computational methods to identify PPARg target genes in the vessel wall.